So many fossils, so little time — to train people to identify them.
As scientists grapple with a vast backlog of marine fossils waiting for identification, an international group led by Yale has begun using machine-learning techniques to tackle the mammoth task facing researchers who study the oceans’ most prolific forms of life.
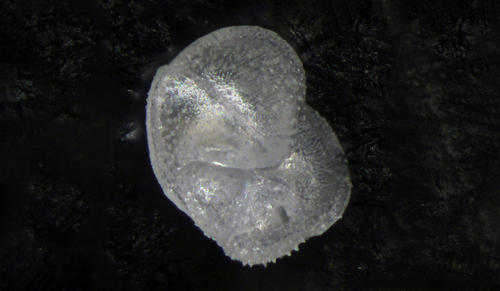
The team, headed by the lab of Yale geologist Pincelli Hull, has built an automated system to wade through vast numbers of plankton fossil images and correctly identify individual species. The new technology represents a major upgrade in scientists’ ability to assess global ecological changes, past and present, via their effect on plankton.
A study announcing the technology appears in the journal Paleoceanography and Paleoclimatology.
“With millions of species on Earth, and many millions more in the fossil record, there are far too few taxonomic experts to identify them all, so that we can understand such critical things as how species and ecosystems respond to climate change,” said Hull, senior author of the study.
“Here we solve this problem by pooling the expertise of taxonomists globally to create the largest database of images, identified to the species level, of an important group of plankton,” Hull added. “We then used machine-learning techniques to train computers to do the same thing.”
Identifying plankton species is central to many areas of ocean paleontology, from conducting geochemical research to understanding the intricate, interconnected dynamics of physical processes in the oceans. They can be analyzed to reconstruct sea surface temperature, salinity, and certain atmospheric values, for example. Yet discerning individual plankton species for research has proven difficult, given the scant resources available to train students in plankton taxonomy.
Hull and her colleagues embarked on an ambitious project to do something about the situation. They compiled a database of more than 34,000 images of typical plankton species, via an online portal called Endless Forams (“forams” is short for foraminifera, which are single-celled organisms with a long fossil record going back hundreds of millions of years) and via a training portal hosted on the citizen science platform Zooniverse. The images came from collections at the Yale Peabody Museum of Natural History and the Natural History Museum in London.
Next, using machine-learning techniques, the researchers trained computer models to identify plankton species. The best-performing model was able to correctly identify 87.4% of the species.
“This is really exciting because it both automates and standardizes an important task,” Hull said. “It increases the repeatability of the science while preserving key knowledge from taxonomic experts.”
The lead author of the study is Allison Hsiang, a former Yale post-doc who is now at the Swedish Museum of Natural History. Co-authors of the study come from institutions in the United Kingdom, Germany, France, the Netherlands, and the United States.
Using supervised machine-learning techniques to answer a biology question presented unique challenges, the researchers noted. Most applications of supervised image classification are used for much different purposes, such as identifying objects in real time for autonomous driving systems or identifying handwritten letters and numbers. Also, certain machine learning techniques for identification, including flipping and rotating the images, can be problematic for identifying taxonomy — and required careful implementation, the researchers said. For example, the identification of some fossils depends on which way its shell is coiled and would change if the image was flipped or rotated.
“Our ultimate goal is to get more data into the hands of the experts,” said co-author Nelson Rios, head of biodiversity informatics and data science at the Yale Peabody Museum of Natural History. “Being able to assess changes in climate over time and understand how species respond is incredibly important.”
Added Hull: “This project has been one of the long-term goals of my research group, and we are delighted to see these results.”